David Wells
My research focuses on using bioinformatics to improve healthcare, with a particular emphasis on infectious disease. At EIT Pathogena I am a senior bioinformatician working on pandemic preparedness and implementing a global pathogen surveillance system. My aims are to improve individual patient care by providing clinicians with the insights from whole genome sequencing; then, provide double benefit from this data by informing public health policy with global pathogen surveillance.
Click for more details of my work...
I co-founded the Quantitative Omics Research group with Dr Michael McAuley. This group aims to bring new mathematical insights to biological data and develop tools to make these analyses accessible.
Previously I worked to prevent infectious diseases through vaccine design and viral genomics at Vaccitech (now Barinthus). During this time I worked on a large number of diseases such as MERS, EBV and its associated cancers, and several autoimmune diseases. Here I developed several tools to aid vaccine design such as HLAfreq and epitope_aligner.
During my research at DIOSVax I worked to develop broadly protective antigens, originally focused on a universal flu vaccine but rapidly focusing on SARS-CoV-2 at the beginning of the pandemic. This project went on to receive $42 million from CEPI. During the pandemic I also partnered with the Royal Papworth hospital as part of the Humoral Immune Correlates of COVID-19 consortium. This project received £1.5 million and one of my roles was managing the results from a battery of separate assays we were performing on all in coming samples.
I use this blog to share interesting results that are too small for a paper, and for demonstrations to get people started with applying some of these techniques to their own data. If you would like to contact me please get in touch through linkedIn.
Posts
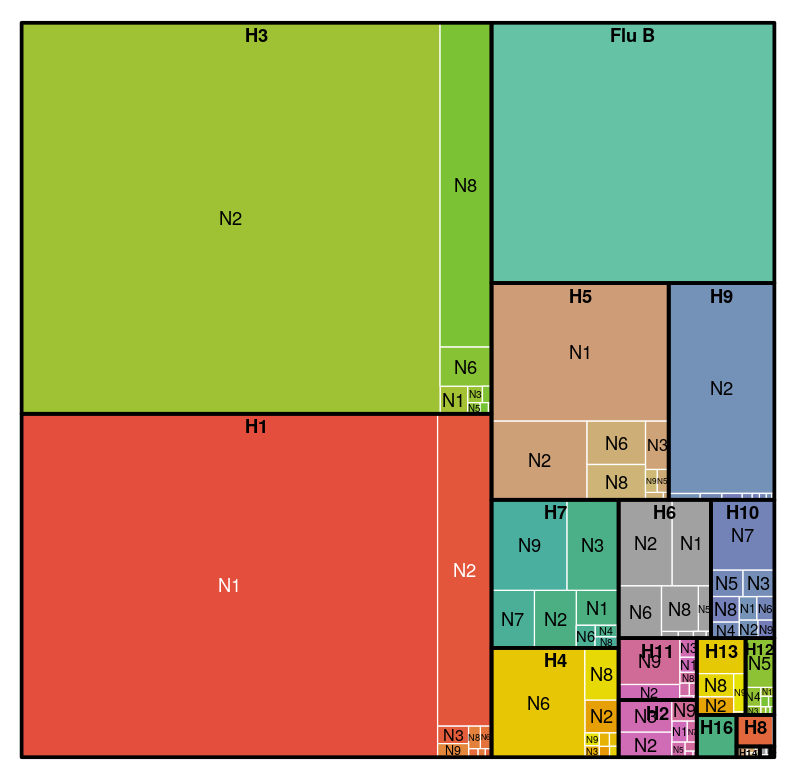
Flu subtypes
How is the flu sequence data distributed across subtypes, hosts and countries on the NCBI data base?
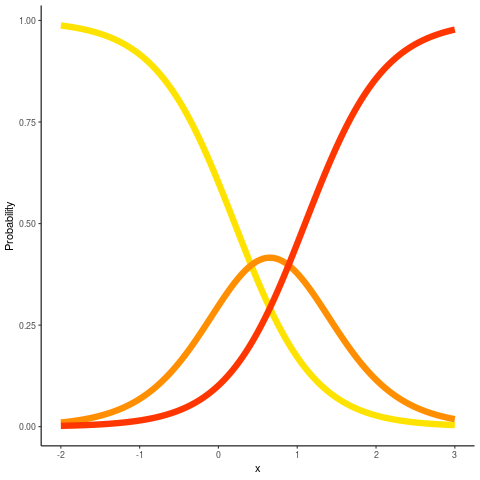
Ordinal GLM
When a response variable is ordered categories you can predict all categories using a single proportional odds model, instead of separate logistic regression models.
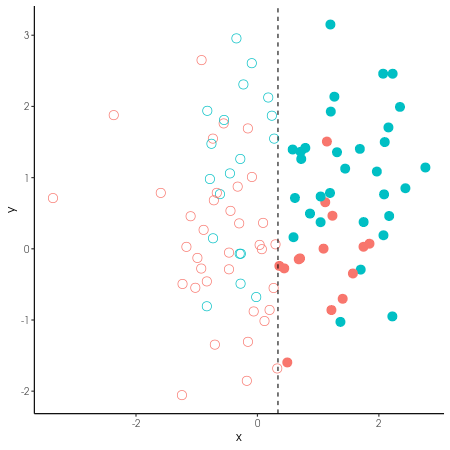
Power & resolution
When binning a continuous variable, e.g. into high and low, positive and negative, you lose information and statistical power.